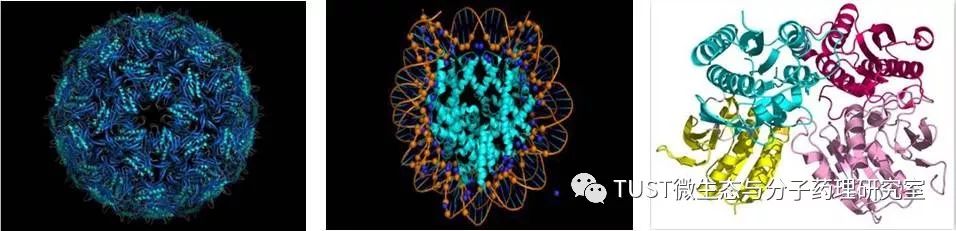
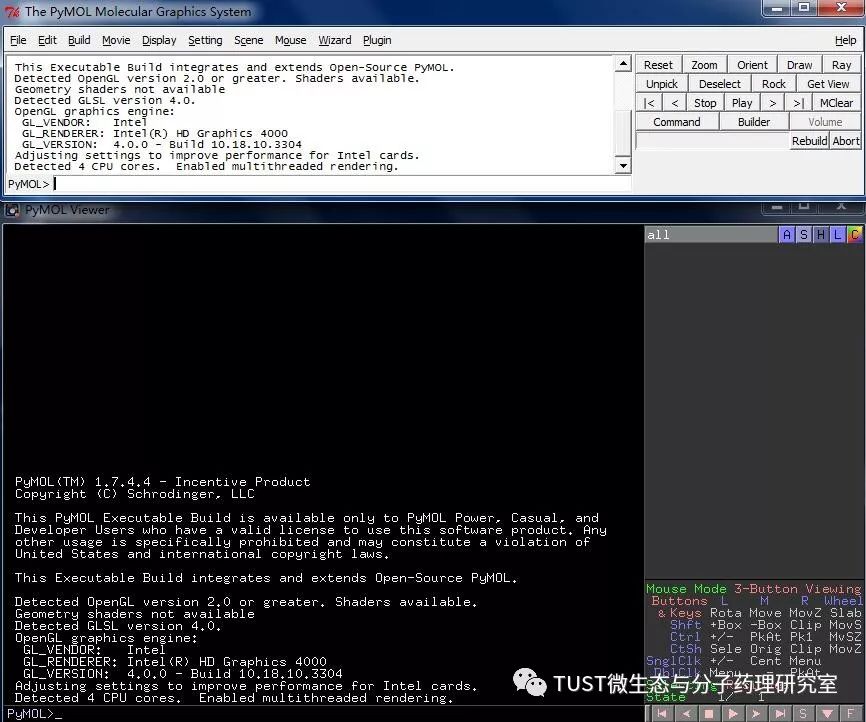
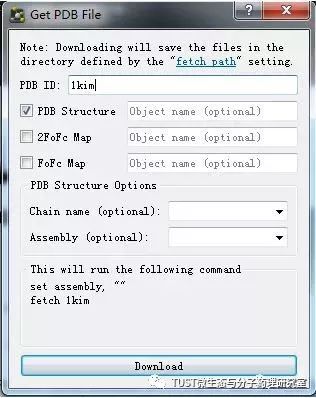
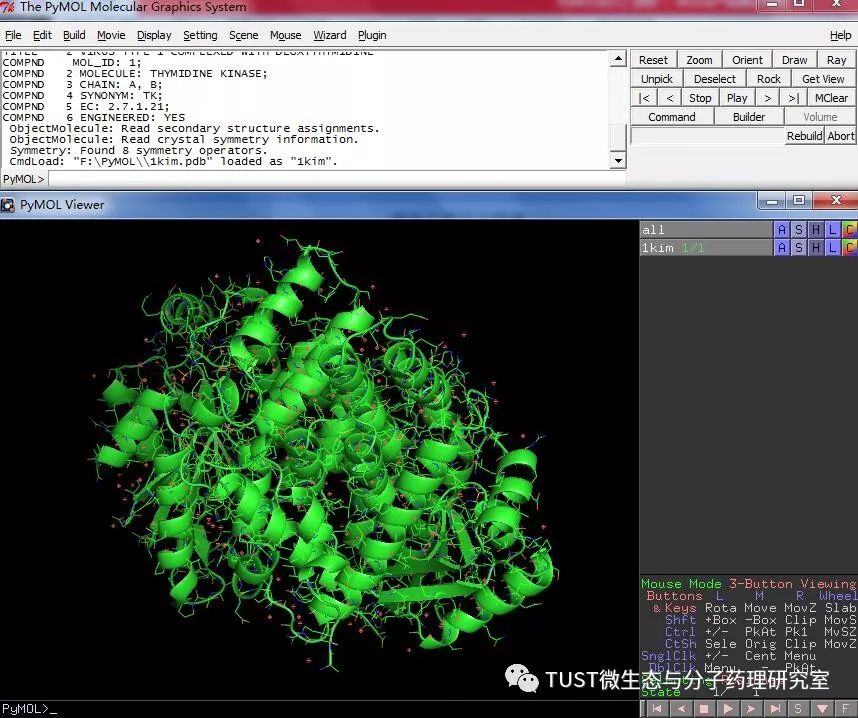
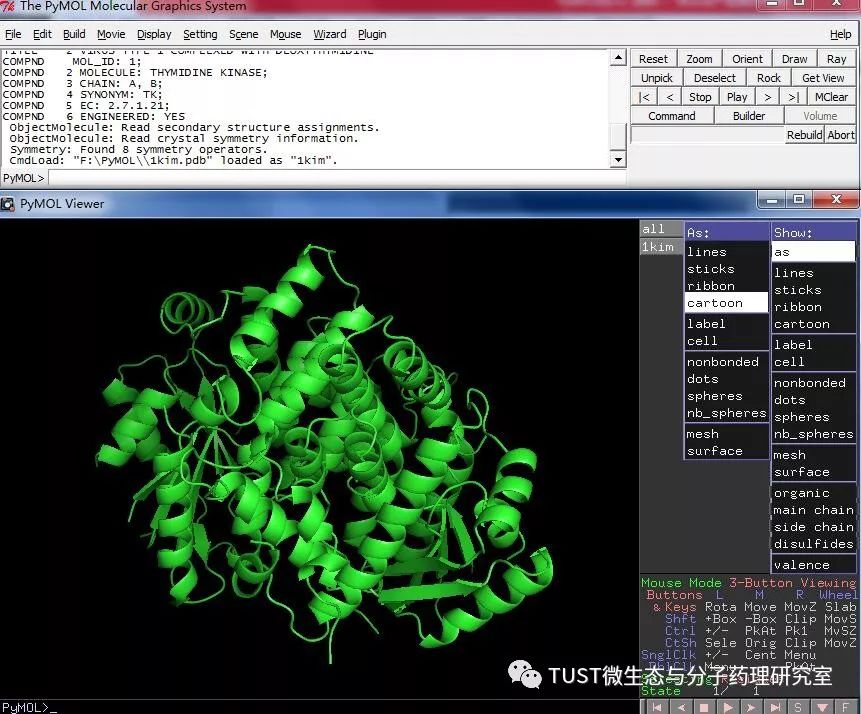
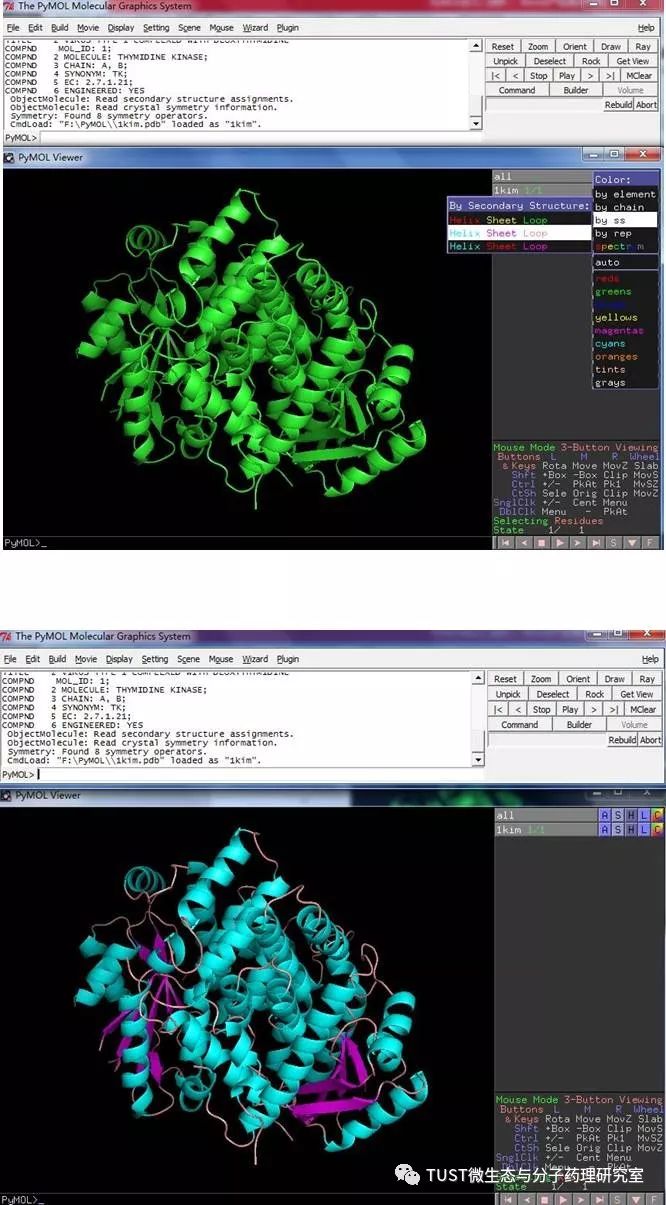
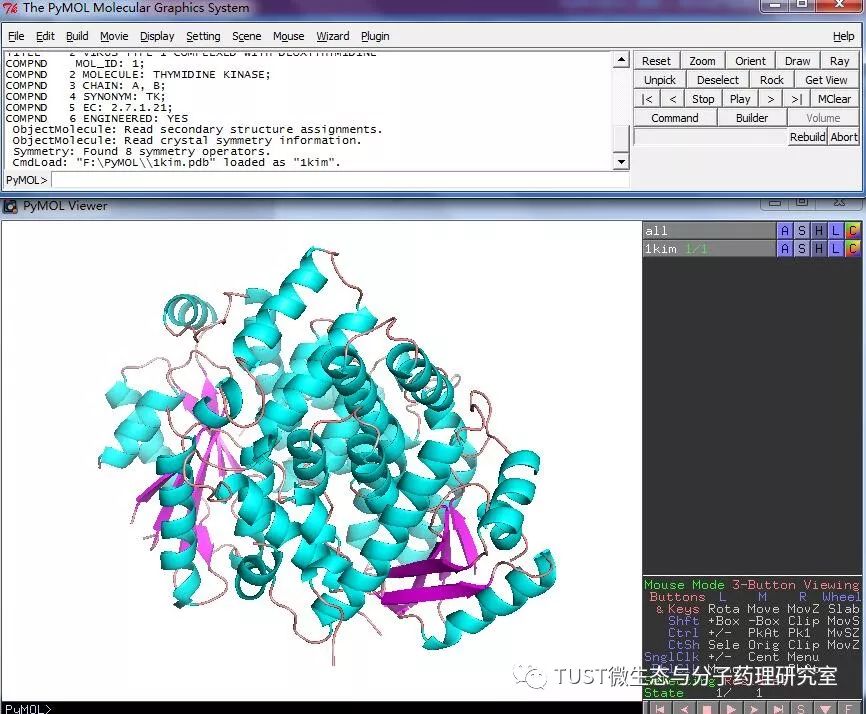
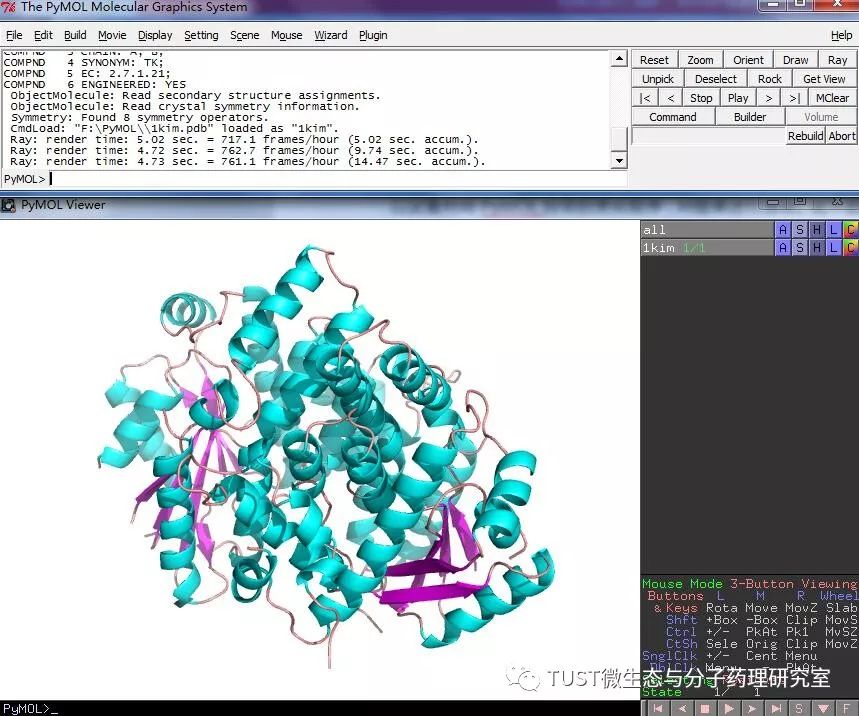
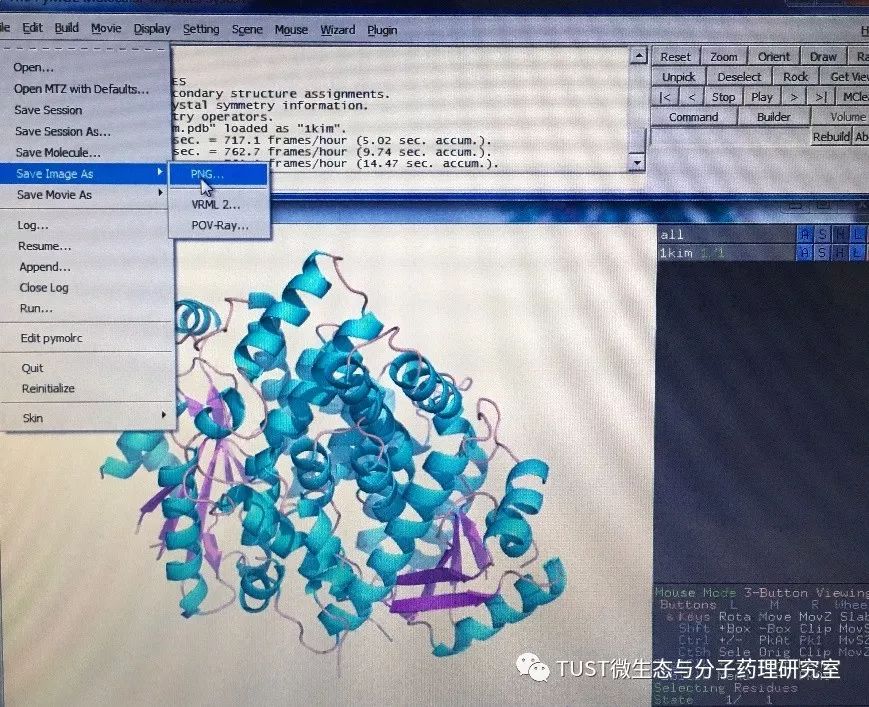
When reading high-scoring literature, do you often find protein structure diagrams that are unforgettable? like this: These pictures can all be made with PyMOL software, so what is PyMOL? The software is named after Py+MOL: "Py" means that it is derived from a computer language Python, and "MOL" means that it is software for displaying the structure of a molecule (English for molecule). It is an open source, written by Warren Lyford DeLano and commercialized by DeLano Scientific LLC. 01 Interface introduction After you open PyMOL, you will see the interface shown below. This interface is divided into 2 windows, which we call the external GUI window (External GUI). We call it the Viewer window. The Viewer window is divided into two parts, the left side is used to display the structure image (viewer), and the right side is an internal GUI window (Internal GUI). The Viewer itself contains a command line (we can see the PyMOL prompt in the lower left corner), which can be used to enter PyMOL commands; in the upper right corner of the Internal GUI we can see five buttons separated by different colors, A , S, H, L, C represent the five words Action, Show, Hide, Label, Color. The corresponding meaning will be demonstrated to you in the following examples. 02 Basic operation The basic format of the protein file is PDB. Take the structure of 1 kim thymidine kinase as an example and explain how the pdb file is displayed. There are three ways to load this file: select File > Open in the External GUI, or use the command line: load Get the following image: Such an image does not clearly show the secondary structure of the protein, and we can make it appear clearer by the following path. Click on S>as>cartoon and the entire protein is shown in alpha helix and beta fold but all are green. At this point, we still feel that they are still not clearly separated. We can distinguish the colors by the following path. Click C>by ss> Helix Sheet Loop, so that different secondary structures can be colored. (SS is secondary structure) The background color of the protein structure that everyone sees in the literature is generally white, but the default background color of PyMOL is black. How do we adjust the background color to white? You can try the following image. Click Display> Background> White in the upper window menu of the External GUI to get the following image. You may find that the resulting image is heavily jagged, so how do you change this? We can try to render the image using PyMOL's own landscaping program. As shown in the figure below, click Draw/Ray in the upper right corner, select Ray, render the image, and directly adjust the image resolution to get the beautified image (the background is transparent, you can directly import the PS to further edit the image). If the configuration of the computer is not enough, we can also choose to use Draw with less system resources and faster speed, and get the following picture. After the adjustment is completed, the image can be saved directly, generally in two forms, one is the PNG image form, and the other is the original PyMOL format, which can be modified later. Oxygen Cylinder,Oxygen Tank,Oxygen Bottle,O2 Tank JIANGSU NEW FIRE FIGHTING TECHNOLOGY CO.,LTD , https://www.newayfire.com